Introduction
The atomistic module is designed to deploy machine learning (ML) models where the atom is the central object. ML potentials and force fields might be the best known cases of atom-centered models. These models are accurate for tasks such as the prediction of energy and atomic forces. They also are powerful because they can generalize to larger molecular as long as the local environments are sampled extensively to cover large domains.
Theory
The basic idea behind atomistic machine learning is to exploit the phenomenon of “locality” and predict molecular or periodic properties as the sum of local contributions:
where \(P_{atom}\) is a functional of atomic positions. In the case the properties are atomic by nature, e.g. atomic forces or atomic charges, there is no need to carry out the sum shown above as the output of the models are atomic.
Atomic Features
ML models require a set of measurable characteristics, properties or information related to the phenomenon that we want to learn. These are known as “features”, and they play a very important role in any ML model. Features need to be relevant, unique, independent, and for our purposes be physics-constrained e.g. rotational and translational invariance.
ML4Chem supports by default Gaussian symmetry functions, and atomic latent features.
Gaussian Symmetry Functions
In 2007, Behler and Parrinello [Behler2007] introduced a fixed-length feature vector, referred also as “symmetry functions” (SF), to generalize the representation of high-dimensional potential energy surfaces with artificial neural networks and overcome the limitations related to the image-centered models. These SF are atomic feature vectors constructed purely from atomic positions, and their main objective is to define the relevant chemical environment of atoms in a material.
For building these features, we need to define a cutoff function (\(f_c\)) to delimit the effective range of interactions within the domain of a central atom.
where \(R_c\) is the cutoff radius (in unit length), and \(r\) is the inter-atomic distance between atoms \(i\) and \(j\). The cutoff function, having Cosine shape, vanishes for inter-atomic separations larger than \(R_c\) whereas it takes a finite value below the cutoff radius. Cutoff functions avoid abrupt changes on feature magnitudes near the boundary by smoothly damping them.
There are different types of SFs to consider when building Behler-Parrinello feature vectors: i) the radial (two-body term) and ii) angular (three-body terms) SFs. The radial SFs account for all interactions of a central atom \(i\) with its nearest neighbors atoms \(j\). It is defined by,
where \(\mathbf{R_{ij}}\) is the euclidean distance between central \(i\) and neighbor \(j\) atoms, \(R_s\) defines the center of the Gaussian, and \(\eta\) is related to its width. Each value in the sum is normalized by the square of the cutoff radius, \(R_c^2\). In practice, one builds a high-dimensional feature vector by choosing different \(\eta\) values.
In addition to the radial SF, it is possible to include triplet many-body interactions within the cutoff radius \(R_c\) through the following angular SFs:
This part of the features is built from considering the Cosine between all possible \(\theta_{ijk}\) angle of a central atom. There exists a variant of \(\mathbf{G_i^3}\) that includes three-body interactions of atoms forming \(180^{\circ}\) inside the cutoff sphere but having an interatomic separation larger than \(Rc\). These SFs account for long-range interactions [Behler2015]:
An atomic Behler-Parrinello feature vector will be composed by a subvector with radial SFs and another subvector of angular SFs. This represents an advantage when it comes to evaluate which type of SFs is more important when predicting energy and atomic forces.
from ml4chem.atomistic.features.gaussian import Gaussian
features = Gaussian(cutoff=6.5, normalized=True, save_preprocessor="features.scaler")
In the code snippet above we are building Gaussian type features using the
ml4chem.atomistic.features.gaussian.Gaussian class. We use a cutoff
radius of \(6.5\) angstrom, we normalized by the squared cutoff raidous,
and the preprocessing is saved to the file features.scaler (by default
the preprocessing used is MinMaxScaler in a range \((-1, 1)\) as
implemented in scikit-learn). The angular symmetry functions used by
default are \(G_i^3\), if you are interested in using \(G_i^4\), then
you need to pass angular_type keyword argument:
features = Gaussian(cutoff=6.5, normalized=True,
save_preprocessor="features.scaler", angular_type="G4")
Atomic Enviroment Vector
The Gaussian feature vectors were modified by Smith et al [Smith2017] in the following way:
The radial symmetry functions are not normalized by the squared of the cutoff radius, and instead of several eta values they use a single value to produce thin Gaussian peaks, and different R_s parameters are used to probe outward from the atomic center.
The angular part adds an arbitrary number of shifts in the angular environment and an exponential factor that allows the angular environment to be considered within radial shells based on the average of the distance from the neighboring atoms.
Atomic Latent Features
Atomic latent features are those extracted using unsupervised learning.
Models
Neural Networks
Neural Network (NN) are models inspired on how the human brain works. They consist of a set of hidden-layers with some nodes (neurons). The most simple NN architecture is the fully-connected case in which each neuron is inter-connected to every other neuron in the previous/next layer, and each connection has its own weight. When an activation function is applied to the output of a neuron, the NN is able to learn non-linearity aspects from the data.
In ML4Chem, a neural network can be instantiated as shown below:
from ml4chem.atomistic.models.neuralnetwork import NeuralNetwork
n = 10
activation = "relu"
nn = NeuralNetwork(hiddenlayers=(n, n), activation=activation)
nn.prepare_model()
Here, we are building a NN with the
ml4chem.atomistic.models.neuralnetwork.NeuralNetwork class with two
hidden-layers composed 10 neurons each, and a ReLu activation function.
Autoencoders
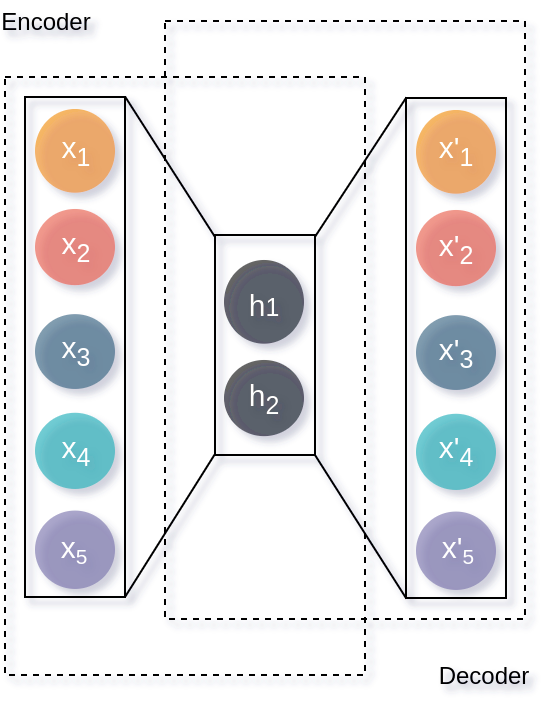
Autoencoders (AE) are NN architectures that able to extract features from data in an unsupervised learning manner. AE learns how to encode information because of a hidden-layer that serves as an informational bottleneck as shown in the figure below. In addition, this latent code is used by the decoder to reconstruct the input data.
from ml4chem.atomistic.models.autoencoders import AutoEncoder
hiddenlayers = {"encoder": (20, 10, 4), "decoder": (4, 10, 20)}
activation = "tanh"
autoencoder = AutoEncoder(hiddenlayers=hiddenlayers, activation=activation)
data_handler.get_unique_element_symbols(images, purpose=purpose)
autoencoder.prepare_model(input_dimension, output_dimension, data=data_handler)
ML4Chem also provides access to variational autoencoders (VAE) [Kingma2013]. These architectures differ from an AE in that the encoder codes a distribution with mean and variance (two vectors with the desired latent space dimension) instead of a single latent vector. Subsequently, this distribution is sampled and used by the decoder to reconstruct the input. This creates a generative model because now we will generate a latent distribution that allows a continuous change from one class to another.
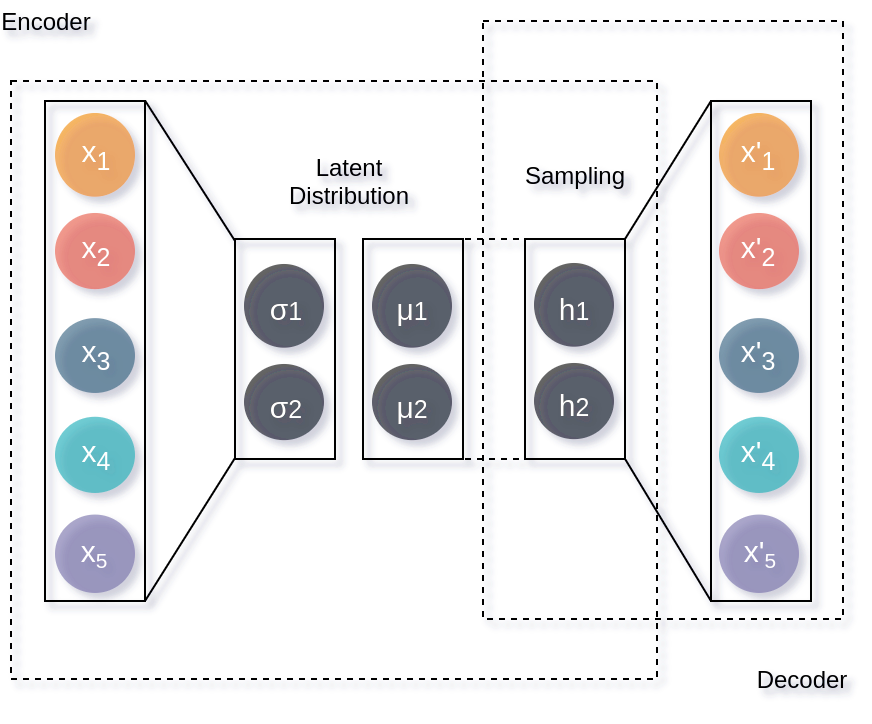
To use this architecture, it just suffices to change the snippet shown above for an AE as follows:
from ml4chem.atomistic.models.autoencoders import VAE
hiddenlayers = {"encoder": (20, 10, 4), "decoder": (4, 10, 20)}
activation = "tanh"
vae = VAE(hiddenlayers=hiddenlayers, activation=activation, variant="multivariate")
data_handler.get_unique_element_symbols(images, purpose=purpose)
vae.prepare_model(input_dimension, output_dimension, data=data_handler)
Kernel Ridge Regression
Kernel Ridge Regression (KRR) is a type of support vector machine model that combines Ridge Regression with the kernel trick. In ML4Chem, this method is implemeted as described by Rupp in Ref. [Rupp2015]. Below there is a description of this implementation:
Molecules are featurized.
A kernel function \(k(x, y)\) is applied to all possible pairs of atoms in the training data to build a covariance matrix, \(\mathbf{K}\).
\(\mathbf{K}\) is decomposed in upper- and lower- triangular matrices using Cholesky decomposition.
Finally, forward- and backward substitution is carried out with desired targets.
Gaussian Process Regression
Gaussian Process Regression (GP) is similar to KRR with the addition of the uncertainty of each prediction.
References:
Behler, J. & Parrinello, M. Generalized Neural-Network Representation of High-Dimensional Potential-Energy Surfaces. Phys. Rev. Lett. 98, 146401 (2007).
Behler, J. Constructing high-dimensional neural network potentials: A tutorial review. Int. J. Quantum Chem. 115, 1032–1050 (2015).
Smith, J. S., Isayev, O. & Roitberg, A. E. ANI-1: an extensible neural network potential with DFT accuracy at force field computational cost. Chem. Sci. 8, 3192–3203 (2017).
Kingma, D. P. & Welling, M. Auto-Encoding Variational Bayes. arXiv Prepr. arXiv1312.6114 (2013).
Rupp, M. Machine learning for quantum mechanics in a nutshell. Int. J. Quantum Chem. 115, 1058–1073 (2015).